Advances in genome technologies have provided unprecedented opportunities for improved understanding of human biology and diseases. Data generation is no longer the major bottleneck of genome medicine. Analysis of high-throughput, genome-wide expression and proteomic data and the integration of genome-wide expression data with clinical outcomes using novel statistical, bioinformational, and computational tools to extract biological information is the newest challenge in trying to make “biological sense” of the massive, complex data sets. With experts in computational biology, biochemistry, and genomics within the Center for Surgery, Innovation & Bioengineering, the Immuno-Metabolic Computational Center (IMCC) tackles medical and biological challenges with novel and innovative solutions.
Read MoreIMCC investigators are working to develop genomics tools and advanced computational methods that could be broadly applicable for use in many different disease studies. Utilizing novel computational approaches to help better understand human systems immunology, IMCC investigators hope to unravel the complex interactions between genes, proteins, metabolites, and different cells and tissues that give rise to the body’s immune-inflammatory and metabolic responses in disease conditions or following injury.
Knowledge based analysis for the interpretation and integration of genomic data
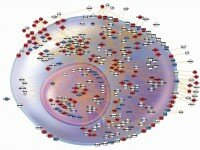
Prototypical inflammatory cell showing composite changes in apparent expression over 24 hr, identifying nodes and interactions
Analysis of high-throughput experimental data alone is not sufficient to identify the underlying biology in healthy individuals, sick patients, or model systems. IMCC investigators developed an approach to systematically examine experimental data in the context of known mammalian biology derived from existing basic and clinical research, to identify pathways and functional modules significant in the disease (in collaboration with Ingenuity Systems, Inc). This approach has been widely applied to many disease studies. In addition, the investigators have developed several methods for statistical analysis of large-scale genomics and proteomics data (in collaboration with Dr. Wing H. Wong from Stanford University and Dr. John Storey from Princeton University), which have been applied successfully to the studies within the “Inflammation and the Host Response to Injury” Glue Grant as well as multiple studies of other diseases.
A current interest of IMCC investigators is to derive disease and tissue specific knowledge bases and use these knowledge bases to better support analyses of genomic data, including prediction of disease outcomes from the data. Using this approach, a knowledge base for the metabolic changes in the human response to inflammation is under development. In related activities, the investigators are developing new approaches for the multi-scale modeling of the disease process by integrating the knowledge base of a particular disease with experimental data.
Computation to translate genome technologies to clinical research
For high-throughput, cost-effective analyses of the human transcriptome in large-scale patient studies, IMCC investigators developed one of the most comprehensive GeneChip microarrays to date (in collaboration with Affymetrix). Compared to RNA-sequencing (RNA-seq), the Human Transcriptome Array (HTA) has been shown to be highly reproducible in estimating expression at both gene and exon levels and reliable in detection of alternative splicing. Bioinformatics and statistical tools for analysis of data from the HTA have been developed by IMCC investigators to enhance the utility of the array, in particular for the analysis of alternative splicing (the ability of one gene to make many proteins). The array platform, which was made commercially available by Affymetrix, has been used in various clinical studies nationwide. The performance of the array was further evaluated as part of the Federal Drug Administration Sequencing Quality Control (SEQC) Consortium, a group that is developing approaches to improve and apply RNA-seq and arrays in combination, for use in large-scale clinical studies.
In collaboration with Dr. Ronald Davis and the Stanford Genome Technology Center, IMCC investigators have led the computational developments in multiple applications of genome technologies to medicine, ranging from original base calling for Next Generation Sequencing (in collaboration with Ion Torrent), sequence analysis of immune repertoire and HLA genes, to the development of stochastic labeling for RNA-seq and cell free DNA analysis (with Cellular Research and Genentech).
Computational studies of the genomic response to inflammation in human diseases and model systems

No qualitative differences in the white blood cell genomic signatures between uncomplicated and complicated patients
Inflammation is a component of the major diseases afflicting the human population, including heart and lung disease, autoimmune diseases, and advanced cancer. The “Inflammation and the Host Response to Injury” Glue Grant program has collected the largest data sets to date on the molecular and clinical outcomes of severe inflammation in patients and in mouse models of inflammation. The computational studies in the Glue Grant reveal that severe acute inflammation in patients leads to an immediate “genomic storm” affecting all major cellular functions and pathways, and the gene responses are highly similar regardless of the different causes of the inflammation. Interestingly, however, the responses for the corresponding mouse models reflect poorly the human conditions upon which they were designed to mimic, and also poorly with one another. Investigators also found that gene signatures can be derived for the prediction of which patients will go on to have complicated clinical outcomes, which are consistent between severe trauma and burns, but cannot be identified from the mouse models.
Given the worldwide prevalence of the use of mice to model human inflammation, these results challenge the assumption that molecular results from mouse models developed to mimic human diseases translate directly to human conditions. The investigators are taking the results from these studies to examine further the genomic response to inflammation in different types of tissues (skeletal muscle and fat) and blood leukocyte cell types (neutrophils, monocytes, T-lymphocytes), including tissue-specific, genome-wide alternative splicing analysis. It is believed that 40-70% of human genes undergo alternative splicing under both normal (healthy) conditions and in disease states.
The novel pathways and modules identified in the Glue Grant patient population may be applicable to the other patient populations in which acute or chronic inflammation underlies the disease. The research interest is to develop innovative approaches to help translate genomic studies of patients to improve the diagnosis, prevention, and therapeutics of various human diseases.
Statistical modeling of the human immune repertoire for disease monitoring
IMCC investigators are very interested in learning more about the “immune repertoire” of an individual, which is the collection of the functional antibodies of the B-lymphocytes and T-lymphocytes in the circulatory system at a given time – a true reflection of the human body’s adaptive immune system. Advances in next generation sequencing enable the high-throughput identification and tracking of individual B-cell or T-cell clones for immune repertoire profiling. Adapting models from population genetics, the investigators developed the statistical approaches to modeling the cellular evolution of the immune repertoire (in collaboration with Stanford University’s Dr. Marcus Feldman), which should provide new knowledge to help investigators distinguish the body’s “disease state” from its “normal” state.
Relevant publications
The Tumor Analysis Best Practices Working Group. Expression profiling—best practices for data generation and interpretation in clinical trials. Nat Rev Genet 2004; 5:229–37
Prokisch H, Scharfe C, Camp DG 2nd, Xiao W, David L, Andreoli C, et al Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2004 Jun;2(6):e160. PubMed PMID: 15208715; PubMed Central PMCID: PMC423137
Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, Baker HV, Xiao W, Inflammation and the Host Response to Injury Large-Scale Collaborative Research Program. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 2005 Mar 29;102(13):4801-6. PubMed PMID: 15781863; PubMed Central PMCID: PMC555033
Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW. Significance analysis of time course microarray experiments. Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12837-42. PubMed PMID: 16141318; PubMed Central PMCID: PMC1201697
Calvano SE, Xiao W, Richards DR, Felciano RM, Baker HV, Cho RJ, Inflamm and Host Response to Injury Large Scale Collab. Res. Program. A network-based analysis of systemic inflammation in humans. Nature. 2005 Oct 13;437(7061):1032-7. PubMed PMID: 16136080
Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Inflammation and the Host Response to Injury Large Scale Collaborative Research Program. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol Cell Proteomics. 2005 May;4(5):700-9. PubMed PMID: 15753121; PubMed Central PMCID: PMC1829297
Liu T, Qian WJ, Gritsenko MA, Xiao W, Moldawer LL, Kaushal A, Inflammation and the Host Response to Injury Large Scale Collaborative Research Program. High dynamic range characterization of the trauma patient plasma proteome. Mol Cell Proteomics. 2006 Oct;5(10):1899-913. PubMed PMID: 16684767; PubMed Central PMCID: PMC1783978
Laudanski K, Miller-Graziano C, Xiao W, Mindrinos MN, Richards DR, De A, et al. Cell-specific expression and pathway analyses reveal alterations in trauma-related human T cell and monocyte pathways. Proc Natl Acad Sci U S A. 2006 Oct 17;103(42):15564-9. PubMed PMID: 17032758; PubMed Central PMCID: PMC1592643
Talasaz AH, Powell AA, Huber DE, Berbee JG, Roh KH, Yu W, et al. Isolating highly enriched populations of circulating epithelial cells and other rare cells from blood using a magnetic sweeper device. Proc Natl Acad Sci U S A. 2009 Mar 10;106(10):3970-5. PubMed PMID: 19234122; PubMed Central PMCID: PMC2645911
Zhou B, Xu W, Herndon D, Tompkins R, Davis R, Xiao W, Inflammation and Host Response to Injury Program. Analysis of factorial time-course microarrays with application to a clinical study of burn injury. Proc Natl Acad Sci U S A. 2010 Jun 1;107(22):9923-8. PubMed PMID: 20479259; PubMed Central PMCID: PMC2890487
Kotz KT, Xiao W, Miller-Graziano C, Qian WJ, Russom A, Warner EA, Inflammation and the Host Response to Injury Collaborative Research Program. Clinical microfluidics for neutrophil genomics and proteomics. Nat Med. 2010 Sep;16(9):1042-7. PubMed PMID: 20802500; PubMed Central PMCID: PMC3136804
Xu W, Seok J, Mindrinos MN, Schweitzer AC, Jiang H, Wilhelmy J, Inflammation and Host Response to Injury Large-Scale Collaborative Research Program. Human transcriptome array for high-throughput clinical studies. Proc Natl Acad Sci U S A. 2011 Mar 1;108(9):3707-12. PubMed PMID: 21317363; PubMed Central PMCID: PMC3048146
Xiao W, Mindrinos MN, Seok J, Cuschieri J, Cuenca AG, Gao H, Inflammation and Host Response to Injury Large-Scale Collaborative Research Program. A genomic storm in critically injured humans. J Exp Med. 2011 Dec 19;208(13):2581-90. Nov 21. PubMed PMID: 22110166; PubMed Central PMCID: PMC3244029 (See also Research Highlight, Nature Reviews Immunology 12, 3)
Logan AC, Gao H, Wang C, Sahaf B, Jones CD, Marshall EL, et al. High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment. Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21194-9. PubMed PMID: 22160699; PubMed Central PMCID: PMC3248502
Seok J, Xu W, Gao H, Davis RW, Xiao W. JETTA: junction and exon toolkits for transcriptome analysis. Bioinformatics. 2012 May 1;28(9):1274-5. PubMed PMID: 22433281; PubMed Central PMCID: PMC3338022
Cuschieri J, Johnson JL, Sperry J, West MA, Moore EE, Minei JP, Inflammation and Host Response to Injury, Large Scale Collaborative Research Program. Benchmarking outcomes in the critically injured trauma patient and the effect of implementing standard operating procedures. Ann Surg. 2012 May;255(5):993-9. PubMed PMID: 22470077; PubMed Central PMCID: PMC3327791
Cuenca AG, Gentile LF, Lopez MC, Ungaro R, Liu H, Xiao W, Inflammation and Host Response to Injury Collaborative Research Program. Development of a genomic metric that can be rapidly used to predict clinical outcome in severely injured trauma patients. Crit Care Med. 2013 May;41(5):1175-85. PubMed PMID: 23388514; PubMed Central PMCID: PMC3652285
Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Inflammation and Host Response to Injury, Large Scale Collaborative Research Program. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013 Feb 26;110(9):3507-12. PubMed PMID: 23401516; PubMed Central PMCID: PMC3587220. (See also Editorial, Nature Medicine, 19, 37)
Finnerty CC, Jeschke MG, Qian WJ, Kaushal A, Xiao W, Liu T, Investigators of the Inflammation and the Host Response Glue Grant. Determination of burn patient outcome by large-scale quantitative discovery proteomics. Crit Care Med, 2013:41:1421-34. PubMed PMID: 23507713; PubMed Central PMCID:PMC3660437
Fu GK, Xu W, Wilhelmy J, Mindrinos MN, Davis RW, Xiao W, Fodor SP. Molecular indexing enables quantitative targeted RNA sequencing and reveals poor efficiencies in standard library preparations. Proc Natl Acad Sci U S A. 2014 Feb 4;111(5):1891-6. . PubMed PMID: 24449890; PubMed Central PMCID: PMC3918775
Ryu SY, Qian WJ, Camp DG, Smith RD, Tompkins RG, Davis RW, Xiao W. Detecting differential protein expression in large-scale population proteomics. Bioinformatics. 2014 Oct;30(19):2741-6. PubMed PMID: 24928210; PubMed Central PMCID: PMC4173009
The SEQC consortium. Power and limitations of RNA-Seq: findings from the SEQC (MAQC-III) consortium. Nature Biotechnology, 2014 (accepted for publication)
Seok J, Xu W, Davis RW, Xiao W, RASA: robust alternative splicing analysis for human transcriptome arrays. Bioinformatics, 2014 (accepted for publication)
Contact
| Wenzhong Xiao, PhD |
| 617-724-7261 |